- Dna Sequence Analysis software, free download. software
- Dna Sequence Analysis software, free download Pdf
- Dna Sequence Analysis software, free download For Windows 10
- Dna Sequence Analysis software, free download Torrent
- Dna Sequence Analysis software, free download Pc
- Dna Sequence Analysis Software
Top 4 Download periodically updates software information of dna sequence analysis full versions from the publishers, but some information may be slightly out-of-date.
View Application Entry for AMAS (Analyse Multiply Aligned Sequences). BVTech Plasmid. BVTech Plasmid is DNA sequence analysis and plasmid drawing software for Windows. It can also be downloaded and installed on local machines.
Using warez version, crack, warez passwords, patches, serial numbers, registration codes, key generator, pirate key, keymaker or keygen for dna sequence analysis license key is illegal. Download links are directly from our mirrors or publisher's website, dna sequence analysis torrent files or shared files from free file sharing and free upload services, including Rapidshare, MegaUpload, YouSendIt, Letitbit, DropSend, MediaMax, HellShare, HotFile, FileServe, LeapFile, MyOtherDrive or MediaFire, are not allowed!
Your computer will be at risk getting infected with spyware, adware, viruses, worms, trojan horses, dialers, etc while you are searching and browsing these illegal sites which distribute a so called keygen, key generator, pirate key, serial number, warez full version or crack for dna sequence analysis. These infections might corrupt your computer installation or breach your privacy. dna sequence analysis keygen or key generator might contain a trojan horse opening a backdoor on your computer.
--- A, B, C ---
AAT - Analysis and Annotation Tool from Michigan Tech University is used to identify genes by comparing cDNA and protein sequence databases.
ABI PRISM software products, from Applied Biosystems, provide advanced, automated analysis for the range of genetic analysis applications from fragment sizing to database matching to pedigree drawing. The complete range of analysis software includes:
- BioLIMS - genetic information management system
- Primer Express - primer design
- DNA Sequence Analysis
- AutoAssembler - sequence assembly
- Sequence Navigator - comparative sequencing
- GeneScan - fragment sizing and quantitation
- Genotyper
- GenBase - storing and sharing sequence information
- GenoPedigree - pedigree drawing
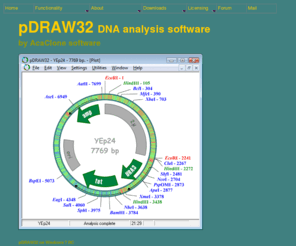
AcaClone pDRAW32 is a freeware DNA cloning, analysis and visualisation software. Also finds DAM and DCM methylations sites, finds and translates open reading frames, calculates optimum PCR annealing temparatures and much more. A detailed description is available at http://www.acaclone.com. Contact: acaclone@hotmail.com
AGCT Hybridization simulation and oligo analysis software especially suited for diagnostic, therapeutic, and common gene family research. New HYBsimulator Mini software, equivalent to OLIGO software at a much lower price. Downloadable HYBsimulator demonstration software for the Power Mac and Windows 95/WinNT. Contact
ALSBYTE Biotechnology Products
PO Box 339
Mill Valley, CA 94942-0339
ALSBYTE Biotechnology Products
2175 E. Francisco Blvd. Suite A4
San Rafael, CA 94901-5524
Tel: 415-457-8502
Fax: 415-457-8541
email: alsbyte1@well.com
Web: http://www.alsbyte.com
AlleleID designs probes and primers for pathogen detection, bacterial identification, species identification and taxa discrimination using qPCR and microarray assays. Align sequences to locate differences in DNA and to find conserved regions. Then proceed to design oligos to amplify and detect only the species or strains of interest from the mix. To design taxa specific or cross species assays, design the least number of oligos to identify a group of genes of interest. AlleleID supports assays for selectively amplifying cDNA from gDNA and designs oligos to detect splice variants using microarrays. Available from:
- PREMIER Biosoft International
- 3786 Corina Way
- Palo Alto, CA 94303-4504
- Tel: 650-856-2703 Fax: 650-618-1773
- Web: http://www.PremierBiosoft.com
Antheprot Protein analysis software. The program is freely available to all academic researchers. ANTHEPROT (ANalyze THE PROTeins) is a package to make protein sequence analysis such as alignment, secondary structure predictions, sites & function detection, physico-chemical profiles, homology search and 3D display of protein structures. This program is now available either for IBM RISC 6000 workstations or IBM PC compatible microcomputers. The main feature of ANTHEPROT is that it is fully interactive within a graphical interface. No particular knowledge about computers is needed and any molecular biologist is able to use it.
Array Designer 4: Design efficient, specific oligos for SNP genotyping and expression microarrays. Start with a list of sequences or the whole genome sequence to study entire organisms effortlessly. Array Designer automatically interprets BLAST results to design specific oligos. Design tiling arrays by avoiding repetitive regions, spot every base of a genomic sequence, to characterize regulatory elements, or to study epigenetic modifications, methylation patterns or protein binding sites. Or design resequencing arrays, far less expensive than the capillary or dideoxy sequencing alternatives, for rapid and accurate populations genotyping studies. Available from PREMIER Biosoft International.
arraySCOUTTM is a gene expression data analysis application that has been developed by LION Bioscience to handle large volumes of data. Key features of arraySCOUT are:
- Interactive graphical visualization
- State of the art gene clustering and statistical analysis
- Integration with external biological databases via SRS
- Project and user management
- Interface to LION's sequence analysis package bioSCOUT®
- Pathway assignment
arraySCOUT is available from
- LION bioscience AG
- Im Neuenheimer Feld 515-517
- 69120 Heidelberg, Germany
- phone: ++49(0)6221/4038-0
- fax: ++49(0)6221/4038-101
- http://www.lionbioscience.com/
Artemis is a free genome viewer and annotation tool that allows visualization of sequence features and the results of analyses within the context of the sequence, and its six-frame translation. Artemis is written in Java , and is available for UNIX, GNU/Linux, BSD, Macintosh and MS Windows systems. It can read complete EMBL and GENBANK database entries or sequence in FASTA or raw format. Extra sequence features can be in EMBL, GENBANK or GFF format. Artemis can be downloaded from the Sanger Web site.
Asterias is a suite of freely- accessible web-based genomic data analysis programs. It is made up of several individual applications for the analysis of genomic data (including microarrays and aCGH data). Most of applications use parallelization (via MPI) and run on a server with 60 CPUs for computation. Thus, the usage of a parallelized-computation server-based application (compared to a desktop or server-based but not parallelized) is readily apparent providing speed ups of factors of up to 50. Most of our applications allow the user to obtain additional information about the genes (chromosomal location, PubMed ids, Gene Ontology terms, etc) by using clickable links in tables and/or figures.
Beacon Designer 7: designs real time PCR assays and supports the following chemistries: TaqMan®, molecular beacons, LNA spiked TaqMan®, MethyLight, Scorpions® primers, NASBA® and FRET assays. Highly specific primers and molecular probes are designed by avoiding cross homologies found by automatically interpreting BLAST search results. Beacon Designer can be used to evaluate pre- designed assays and designing compatible oligos to extend the designs. It can design molecular beacons and TaqMan® probes for single tube multiplex experiments and TaqMan® probes for identifying DNA methylation patterns. Available from PREMIER Biosoft International.
Bio Image is a life sciences software information company which carries a wide variety of electrophoresis image analysis software for Windows, Powermac, and UNIX. Software and hardware solutions include 2-dimensional electrophoresis, 1-dimensional electrophoresis (with special products for RFLP, RAPD, phylogenetic studies, and other genetic mapping applications), High Density Grid, DOt and Slot Blot, and Colony Counting. Contact:
Bio Image
525 Avis Drive, Suite 10
Ann Arbor, MI 48108
Tel: 313-930-9900
Fax: 313-930-0990
Email: info@bioimage.com
URL: http://www.bioimage.com
BioinformatiX is an enterprise software which provides an environment for the analysis of microarray data. It is available from:
- Xpogen
- 80 Prospect Street
- Cambridge, MA 02139
- Tel: 617-386-4302
- http://www.xpogen.com
BioRainbow Analysis Tools are a collection of software tools for binding site prediction, weight matrix search, regulatory sequences analysis, microarray analysis, footprint. Most of tools a freeware or have a free trial and avaliable for download. Most of tools are for Win32. If you require a specific tool - you can request for it. For more information, check the BioRainbow Tools web site.
bioSCOUT® is a comprehensive and customizable bioinformatics package. It is a turn key system which guides the user automatically through all processes of gene sequence analysis. It makes use of the world's strongest data integration system, SRS. bioSCOUT can either be installed on a central company server and accessed by multiple users via intranet, or clients can optionally access the high performance servers at LION Bioscience via a secure data line connection.
bioSCOUT is available from
- LION bioscience AG
- Im Neuenheimer Feld 515-517
- 69120 Heidelberg, Germany
- phone: ++49(0)6221/4038-0
- fax: ++49(0)6221/4038-101
- http://www.lionbioscience.com/
BioTools offers three primary bioinformatics products: GeneTool for DNA sequence analysis, PepTool for protein sequence analysis, and ChromaTool for chromatogram analysis. These products offer scientists a single environment in which to rapidly search public and proprietary databases, probe sequence homologies, access functional information, align multiple sequence, predict the secondary structure of proteins, and provide a single, powerful and intuitive interface to access sequence data and analytical tools. All three products provide a unified user interface on a number of platforms including Sun Solaris, SGI, LINUX, Windows, and PowerMac; providing a comprehensive sequence analysis suite.
Contact:
- BioTools Incorporated
- 800 Ironwood Professional Centre
- 10050 - 112 Street
- Edmonton, AB
- Canada
- T5K 2J1
- web: http://www.biotools.com
- email: biotools@biotools.com
BlockSearch is a quantitative method for the elucidation of unknown protein functions. The program is written by Rainer Fuchs, GlaxoWellcome, in C and runs on UNIX systems. It is available as program number 668 from QCPE
Bosque (http://bosque.udec.cl) is a distributed software environment oriented to manage the computational resources involved in typical phylogenetic analyses. Bosque has been implemented as a client-server application where the server can execute installed phylogenetic programs (Phylip, PhyML, TreePuzzle, Muscle) and the client manages the results on a local relational database, although it can also execute phylogenetic programs locally, useful when no server is available. The client also performs the graphical visualisation and edition of trees and alignments, providing an environment for the analyses, from the integration of sequences to the printing of a final tree.
Clann: Software for investigating phylogenomic information using supertrees. The word 'Clann' is the Irish word for 'family'. This version of Clann (2.0.1) implements four supertree methods (which are used as optimality criteria):
- Matrix Representation using Parsimony (But Paup* is necessary for the parsimony part)
- Maximum Quartet fit
- Maximum Split (component) fit
- Most Similar Supertree Method (A novel method related to the average consensus method with branch lengths set to unity)
For each of these optimality criteria, Clann allows:
- Searches of supertree-space using Exhaustive or Heuristic searches (NNI & SPR)
- Searches of User-defined trees.
- Investigation into the support for various hypotheses through:
- Bootstrapping (at the source tree level)
- YAPTP tests (a randomisation test similar to the ptp test)
Clann is freely available to the scientific community at: http://bioinf.may.ie/software/clann. Executables are available for Linux, Mac and Windows.
CLC Free Workbench is a freeware bioinformatics software package available for Windows, MacOS X and Linux.
CURVES, by Richard Lavery and Heinz Sklenar is a very useful nucleic acid helical analysis program. The current version is CURVES 5.1 and its available free directly from the author. Contact:
Richard Lavery
Laboratoire de Biochimie Theorique
Institut de Biologie Physico-Chimique
13, rue Pierre et Marie Curie
75005 Paris, France
Tel: +33 1 43 25 26 09
Fax: +33 1 43 29 56 45
Email: richard@ibpc.fr
--- D, E, F ---
DNADynamois a general purpose software for DNA and Protein sequence analysis, available for Windows and OSX. DNADynamo can be used to align abi/scf trace sequencing files, restriction site analysis, sequence annotation, oligo design and storage, virtual sub-cloning, plasmid maps, BLAST search, multiple alignments and much more. A free trial version is available from http://www.bluetractorsoftware.co.uk.
DNASIS is a robust sequence analysis software package that delivers industry standard functionality - molecular biology tools, multiple consensus calling methods, and rapid BLAST searches - as well as unique features that enhance common task performance and allow user-defined product customization using the product's unique DNASpace automation component and open application programming interface (API). Program runs on Microsoft Windows® 98, NT 4.0 (SP6 or later), 2000, XP. For additional information, contact:
- MiraiBio
- 1201 Harbor Bay Parkway, Suite 150
- Alameda, CA 94502
- Tel: 800-624-6176
- Web Site: http://www.miraibio.com
DNPTrapper is a shotgun sequencing assembly editing tool, specifically designed for finishing and analysis of repeated regions. The DNPTrapper source code (it is Open Source) can be downloaded athttp://dnptrapper.sourceforge.net. The paper describing the software is located in BMC Bioinformatics.
EuGene and SAm is a menus based DNA and protein sequence analysis package. The program efficiently manages data during DNA sequencing projects. The system can simulate genetic engineering experiments, search databases of DNA and protein sequences, compare multiple sequences for similarities and provide detailed structural information of sequences. Runs on SUN workstations. Contact:
Christine B. Powaser
Lark Sequencing Technologies
9545 Katy Freeway, Ste 200
Houston, TX 77024 USA
Tel: 713-464-7488
Fax: 713-464-7492
Expression is a powerful application for DNA and protein sequence analysis. Utilising a novel interface, Expression makes complex computational analyses of sequence information incredibly simple. Expression uses the very latest computing technology to set new standards in the way sequences are analysed. Features include: Sequence Annotation, Interactive Graphical Sequence Map, Degenerate DNA and Amino Acid Sequence Support, Restriction Analysis, PCR Primer Design and Analysis, ORF Prediction, Pattern Finding, Reverse Translation, GenBank Searching, Pattern and Motif Identification, Multiple Sequence Alignment, and Protein Structure Prediction. The graphical representation of annotated sequences allows the user to create publication quality diagrams of plasmids, genetic constructs, etc. The lightning fast algorithms used in Expression allow virtually all computations to occur in real-time, greatly simplifying the analysis approach. The intuitive interface amd comprehensive set of online tutorials means that Expression is easy to learn for professionals in the laboratory, as well as being readily adaptable for educational purposes. Expression runs on Microsoft Windows® 95/98/NT/2000/XP. For additional information, check the Genamics web site.
--- G, H, I ---
Genchek, developed by Ocimum Biosolutions is a comprehensive, LIMS based, user friendly Nucleotide and Polypeptide Sequence Analysis Tool with a backend Relational Database. With Genchek you can design Primers, annotate sequences, view annotations with publication quality graphics, Import/ Export and modify chromatograms, perform BLAST searches, SNP search on sequences and Contig assembly, find Genes with proprietary Neural Network algorithms, analyze Restriction Enzyme sites, perform a Six Frame analysis and find ORFs of sequences, calculate hydrophobicity plots for Polypeptide Sequences and manage all this information among different users with different privileges on a backend relational database. For further details or demo email at genchek@ocimumbio.com Genchek 2.0 is a Mac OSX, Windows, Linux and Unix compatible program.
- Ocimum Biosolutions
2915 Redhill Avenue, Suite F202
Costa Mesa, CA 92626
Tel/Fax: 866-227-7512 (Toll Free in US)
- Ocimum Biosolutions LLC
- Fortune Park VI
- 8765 Guion Road, Suite G
- Indianapolis, IN, 46268
- Tel: 1-317-228-0600
- Fax: 1-317-228-0700
- 1-866-227-7512 (Toll free)
- Web site: http://www.ocimumbio.com/web/d efault.asp
Genehound(™) offers a new, innovative, and exciting apporach to identifying coding regions in prokaryotic genomes. Preliminary results indicate that it may be the most accurate gene identification program developed to date. It is easily trainable for virtually any prokaryotic genome. It readily identifies protein coding regions and the corresponding reading frame, as well as start and stop codons. GeneHound(™) operates with a graphical user interface, making it easy to use. Supported platforms are Windows 95/98/NT standalones. For more information, visit our website at http://www.apocom.com or contact ApoCom at 423-927-6120 or apocom@apocom.com.
GeneInform is an easy-to-operate gene expression management and analysis tool that saves cost and time by facilitating the collection, storage, analysis, and sharing of gene expression data. Gene expression analyses require large numbers of experiments and volumes of data. Organizing this data for efficient access, analysis, and collaboration is a challenge. GeneInform addresses many of the problems found in expression analysis.
- Gene Networks
- 560 South Winchester Blvd.-Suite 500
- San Jose, CA 95128
- Tel: 408-572-5567
- Web: http://www.gene- networks.com
Gene Inspector(™)1.5: A powerful and versatile combination of an electronic laboratory notebook and sequence analysis package for biologists. GI 1.5 delivers a wide range of enhancements to the award- winning Gene Inspector 1.0, including extensive drag and drop options, more multiple sequence alignment match and display options, and increased speed in its routines. There are over 640 different ways to display multiple sequence alignments to highlight exactly what the user wants to demonstrate. With GI 1.5, users can select GI sequences to run SIGNALP protein searches, as well as BLAST, BLOCKS, FASTA, and GRAIL nucleic acid and protein searches over the Internet.
To learn more and to download a demo of Gene Inspector 1.5 (Macintosh only) visithttp://www.sciquest.com. Contact
- SciQuest, Inc.
- P.O. Box 12156
- Research Triangle Park, NC 27709-2156
- phone: 1-877-710-0412
- fax 1-888-638-7934
GeneJockey is a program for editing, manipulation, and analysis of nucleic acid and protein sequences. It can scan nucleic acid sequences to select oligonucleotides for use as PCR primers. It can also perform sequence alignments and concatenation and can search GeneBank and other libraries. Available from Biosoft, PO Box 10938, Ferguson, MO 63135; Tel: 314-524-8029; URL http://www.biosoft.com/biosoft
GeneLinker products are the easiest way for researchers to start analyzing gene expression data. Within ten minutes of installing the software, novice users are able to start exploring their data and learning things that would be impossible with tools like Excel, or would take significant training and time with our competitor's products. The GeneLinker Platinum product also provide advanced analysis features that aren't available in any of our competitor's offerings, making one of the GeneLinker products the perfect choice for any lab. Features support:
- Data Import - All of the GeneLinker products provide an easy to use data import process that supports text files, 2 color data and a variety of native chip for
- Normalization - GeneLinker offers a variety of normalization options, including lognormal scaling, scaling to a baseline sample, linear ratio-based methods, normalizations using control genes, or lowess.
- Filtering - GeneLinker provides a number of ways of prioritizing genes, including statistical prioritization, profile matching, N-fold induction and repression, Gene Lists and a variety of others.
- Clustering - GeneLinker allows partitional clustering of genes or samples (or both) using Self-Organizing Maps (SOMs), K-Means and mutual nearest neighbors (Jarvis- Patrick) methods and hierarchical clustering of genes or samples (or both) using single, average or complete linkage.
- Statistics - GeneLinker allows you to find out which genes vary most significantly across a range of conditions with F-test and Kruskal-Wallis tests.
- Visualization - GeneLinker supports all of the visualizations that you require for Gene Expression Analysis including: Scatter Plots, a variety of 2D and 3D Score Plots, Histograms, Matrix Plots (Colour Array Plots), Matrix Tree Plots (1 and 2 way Dendrograms), SOM Plot, Profile Plots and many others.
- Exporting Graphics - GeneLinker allows you to export graphics for the web in PNG format or publication-quality graphics using either SVG or PDF formats.
- Reports - The experiment navigator provides a detailed audit trail should you wish to repeat an analysis or an entire workflow.
- Performance - All of GeneLinker's analysis methods have been optimized for speed and memory use on large data sets. They are all implemented in C++ giving them a tremendous advantage compared with products implemented entirely in Java.
- Web Links - You can select genes in visualizations and look them up in external web databases such as GenBank, Unigene, and NetAffx.
- Predictive Patterns Software
- 188 Westdale Avenue
- Kingston, ON Canada :: K7L 4S5
- Tel: (613) 483-9803 or (613) 539-1126
- Fax: (613) 353-1772
- Website: http://www.predictivepatterns.com
GENEMARK is a genefinding tool available from the Georgia Institute of Technology that uses an algorithm based on non-homogenous Markov chain models.
GENEPARSERis a coding region recognition program from the University of Colorado that uses potential similarity between query sequence and known amino acid sequences.
GeneSifter™, a Web-based microarray analysis system that combines data management and analytical functions with integrated, current gene annotation from databases such as Unigene and LocusLink. The product's Web-based design complements traditional applications while providing users with the convenience of anytime, anyplace, any platform access. It smoothly facilitates collaboration throughout an organization or across the world. Thirty day free trial available by signing up at the GeneSifter website.
Contact:
- Vizx Labs, LLC
- Suite 300
- 2815 Eastlake Ave East
- Seattle Wa 98102
- Tel: 206-336-5549
GeneSolve is a single-User desktop sofware package for analyzing nucleic acid sequence infromation. The program runs on Microsoft Windows 98, NT, 2000; PowerMac, Solaris 2.6, Irix 6.2+ and Linux-x86. For additional information,
- Stratagene
- 11011 North Torrey Pines Road
- La Jolla, CA 92037 USA
- Tel: (800) 424-5444 or 800-894-1304
- Fax: (512) 321-3128
- Web: http://www.stratagene.com
GeneStudio Pro from GeneStudio, Inc. (http://www.genestudio.com) is a newly developed suite of molecular biology programs for Windows. Features include:
- Modern, intuitive, drag-and-drop interface
- Sequence alignments with an interface to phylogenetic analysis and display of trees
- Contig editor for your sequencing projects
- Innovative viewers interfacing with word processing and presentation software
- Full integration of Internet resources such as Entrez (GenBank), PubMed, and BLAST
GeneWorks - an integrated sequence analysis and database searching on the Macintosh previously marketed by Oxford Molecular Group. The program is no longer available.
Genie, from LBNL, is a gene finder based on generalized hidden Markov models to locate multi-exon genes.
GENLANG from the University of Pennsylvania, is a syntactic pattern recognition system that uses tools of computational linguistics to locate genes.
GenomeBrowser is a powerful software tool that simplifies the proccess of analysis, annotation, and manipulation of genetic sequences. The underlying principle of UniPro GenomeBrowser is an interactive visualization of genetic sequences with ability to navigate, zoom and create annotations for selected parts of sequences. Advanced search tool, powerful contextual statistics, support of major genetic data formats and collection of pluggable modules increase the efficiency, simplify and, in a number of cases, make the process of analysis automatic. Final results as well as intermediate ones are graphically presented and can be saved and used in a future analysis.
Genowiz, developed by Ocimum Biosolutions is a powerful gene expression analysis program designed to efficiently store, process and visualize gene expression data. It includes a suite of advanced analysis methods and offers choice for selecting analysis that is appropriate to a researcher's dataset. Genowiz™ provides numerous visualization options to track down intricate correlation in microarray data. Genowiz allows researchers to organize experimental information, quickly and easily import data and image files, preprocess, normalize, plot, perform cluster analysis, classify, visualize patterns, review gene information, as well as link analysis results to external tools. For further details or demo email at genowiz@ocimumbio.com. Genowiz is a Mac OSX, Windows, Linux and Unix compatible program. Additional infromation can be obtained from:
- Ocimum Biosolutions
2915 Redhill Avenue, Suite F202
Costa Mesa, CA 92626
Tel/Fax: 866-227-7512 (Toll Free in US)
GENSEARCH is a user-friendly locus specific software application assisting biologists in the analysis of DNA sequences. Gensearch quickly and reliably proposes potential mutation locations, interpreting their potential consequences, and offering all necessary information when evaluating his DNA sequence data. Gensearch has been validated on the LDLR gene with more than 300 patients, the equivalent of around 6000 amplicons, demonstrating its high specificity and selectivity as well as an impressive gain in time versus the current semi-manual process. A database module is under development. For additional information,
- PhenoSystems SA
- Rue Raymond Lebleux 114
- B-1428 Lillois Witterzee
- Belgium
- Tel: +32 2 732 57 93
- Fax: +32 2 734 77 40
- http://www.phenosystems.com
GLIMMER from John Hopkins, is a program that uses interpolated Markov models to find genes in microbial DNA.
GoCore V is a free, open source, 'protein sequence alignment and analysis tool that operates as an Excel add-in. It is compatible with Excel 98 or above and Windows 95 or above. A Mac OS X version is also currently under development and expected to be released before the summer.
As well as containing many standard protein sequence analysis tools, GoCore is particularly effective at combining the different kinds of information obtained from cross-species alignments and protein family alignments to make useful functional predictions. Full details, online documentation and free download are available.
Grail(™) Toolkit. The newest version of Grail (tm), ApoCom Grail Toolkit has improved accuracy and capabilities, and continues to be one of the best tools on the market for identifying genes and proteins in the human genome. This superior gene identification and characterization software runs on Grail EXP, the latest algorithm for Grail. It's features include homology-based gene assembly, dynamic memory allocation, and complementary modes. The most exciting feature is its new dynamic memory allocation, which allows you to use a data string of unlimited size and complexity. Running with a Java-based interface, using Grail is simply point and click. A command line interface allows you to channel the output directly into your proprietary data pipleine. Supported network platforms include Unix (DEC Alpha and IBM RS6K), Silicon Graphics, and SUN. Supporte client platforms include Unix, Mac, and Windows 95/98/NT. For more information and an online demo, visit the ApoCom website at http://www.apocom.com or contact them at 423-927-6120 or apocom@apocom.com.
Genomix GrailEXP6 is created by the authors of the Grail, GrailPro, and GrailEXP3 systems. Those same authors founded, operate, and continue cutting edge research at Genomix Corporation. EXP6 is offered exclusively by Genomix Corporation and is three full generations beyond any other Grail/EXP systems available through academic or commercial sources. Genomix EXP6 is regarded by many as the best package in the world for recognizing splice sites, identifying exon/intron boundaries, and modeling genes. GenePool from Genomix provides the entire human genome with comprehensive splice variants and unsurpassed exon accuracy processed using the EXP6 algorithm. All results are backed with experimental transcript information making GenePool the best representation of the human genome available.
Genomix Corporation is a genomics based drug discovery company committed to generating, providing, and characterizing novel gene and protein candidates for the treatment of many different types of human disease. Genomix is using its unique technologies to generate a holistic view of genes and alternatively spliced gene products implicated in human disease. Many gene-based human diseases are caused not by the known and expected protein from the disease-related gene, but from alternative proteins produced by the gene that are specific to the disease condition and tissue type. Genomix has computationally characterized over 80,000 transcripts in the human genome and developed a portfolio of novel disease-implicated gene and protein candidates. This growing catalog includes novel members of virtually all classes of pharmaceutically important protein targets such as cytokines, chemokines, nuclear receptors, and signaling proteins. This catalog provides significant potential for the development of new and powerful tissue and disease specific therapies and diagnostics based on such alternative genes and proteins.
Contact:
- Genomix Corporation
- 1020 Commerce Park Drive
- Oak Ridge, TN 37830
- Phone: 865-220-0043
- Fax: 865-220-0053
- URL: http://www.genomix.com
Iditis - This package provided instant access to protein motifs, substructures and interactions buried in the Brookhaven databank. Iditis Architect is a suite of programs that lets you create protein structure data for inclusion in the Iditis database. The program was developed by Oxford Molecular Group, but it is no longer available.
ISYS(TM), is the National Center for Genome Resources' new product that integrates independent bioinformatic software tools and databases. This software tool enables genomic investigators to go beyond single-focus analysis tools by creating an integrated environment in which they can more easily and rapidly discover new knowledge. ISYS is capable of integrating data sources and analysis tools from an investigator's own laboratory with those developed elsewhere. Free evaluation copies of ISYS can be downloaded and installed from the ISYS Web site, http://www.ncgr.org/isys.
ISYS's key technical strength is the platform's ability to couple separately developed Java(TM) software components and analysis tools, harnessing their collective strength while allowing them to evolve independently. ISYS uses DynamicDiscovery(TM) technology to allow users to find appropriate paths from one software component to another, according to their particular data sets and configuration of components. DynamicDiscovery is used to integrate Java programs and to allow the integration of Web pages with those programs. ISYS's Web integration feature works generically with virtually any Web page, or it can be augmented with even richer functionality if pages are specially 'marked up' for integration with the system. ISYS has the ability to mark up several popular bioinformatics Web sites and lets users of the system extend it to mark up their own favorite pages. In addition, ISYS has a published API (Application Programming Interface) to allow users to adapt their own programs and databases for integration with the platform. Alternatively, NCGR developers and scientists will work with organizations, one-on-one, to tailor ISYS to meet their specific discovery needs.
--- J, K, L ---
JAligner is an open source Java implementation of the dynamic programming algorithm Smith-Waterman for biological pairwise local sequence alignment. Please find below a summary of the features of Jaligner:
- Affine gap penalty (open and extend) is supported, however, the overall time complexity of the alignment process is maintained at O(n2) -as the orignial Smith-Waterman algorithm- by using Gotoh's improvement for the computation of the penalties.
- The space complexity for calculating and holding the similarity scores is reduced from O(n2) to O(n) by using a single-dimensional array of size n instead of the original two-dimensional similarity array of size n*n.
- The two-dimensional array of size n*n, for holding the directions (Diagonal, Left, Up and Stop) for the trace back step, is mapped into a single-dimensional array of size n*n. This mapping approach significantly speeds up the processing time of allocating the required memory space for the array because the Java Virtual Machine (JVM) simply allocates an array of n*n floats (Primitive Data Type), instead of allocating an array of n 'objects', each of which is an 'array' of n floats.
- There are available 70 scoring matrices, that were collected from the NCBI FTP site. Additionally, it is possible to use user-defined scoring matrices.
- Usage is through the command line, as a desktop application or as a Java Network Launch Protocol (JNLP)-based application with a simple and friendly Graphical User Interface (GUI).
Lasergene: Sequence Analysis software for Windows and Macintosh computers. LASERGENE is modular, for single users or networks, offering in-depth coverage of sequencing, primer design, sequence alignment, databases and database searching, protein analysis, and restriction map analysis. Modules include:
- PRIMERSELECT for Primer and Probe Design and Analysis
- PROTEAN for Protein Sequence Analysis
- MEGALIGN for Multiple and Paired Sequence Alignment
- MAPDRAW for Restriction Analysis and Maps
- SEQMAN for Sequencing
- GENEMAN for Databases and Database Acquisition
- EDITSEQ for Sequence Editing
LASERGENE is available from DNASTAR Inc. at:
- 1228 South Park Street
Madison, Wisconsin 53715 USA
Tel: 608-258-7420
Fax: 608-258-7439
Email: sales@dnastar.com
URL: http://www.dnastar.com - GATC GmbH
Fritz-Arnold-Str. 23
D-78467 Konstanz
Germany
Tel: ( 49) 7531 57209
Fax: ( 49) 7531 57313
Email: gatc@lakelink.cl.sub.de
Look: is a set of informatics tools which allows the researcher to seamlessly access and manage protein sequences, structures, literature and experimental data. The product has been retired and is no longer available.
--- M, N, O ---
MacImdad was macromolecular modeling software for the Macintosh from Molecular Applications Group. The program is no longer available.
MacVector is a set of sequence tools which conduct DNA sequence analysis using Macintosh. Available fromAccelrys.
MORGAN, from John Hopkins, is a decision tree program for finding genes in vertebrate DNA.
MPSRCH: is a program which permits rapid searches of DNA and protein sequence data banks using the full Smith-Waterman algorithm for the most rigorous searching available. The program is available fromEdinburgh Biocomputing Systems Limited. The authors recently have ported MPSRCH to general purpose hardware. It can be accessed free of charge at the company's website.
NeoBio is a Java class library of computational biology algorithms. The current version consists mainly of pairwise sequence alignment algorithms such as the classical dynamic programming methods of Needleman & Wunsch (global alignment) and Smith & Waterman (local alignment). A more efficient approach, due to M. Crochemore, G. Landau and M. Ziv-Ukelson (2002) is also available.
All sequence alignment algorithms support simple scoring schemes as well as substitution matrices such as standard BLOSUM and PAM matrices with constant (linear) gap penalty functions only. Future versions may contain related algorithms such as multiple sequence alignment, database search and protein structure prediction. NeoBio also also provides simple GUI and command line based tools to run the sequence alignment algorithms on DNA and protein sequences.
NetPrimer: Performs and prints individual primer and pair analysis using the latest primer design software. Reputed to be the most comprehensive free primer analysis program on the web. Available from
PREMIER Biosoft International
3786 Corina Way
Palo Alto, CA 94303-4504
Phone: 650-856-2703
Fax: 650-843-1250
http://www.PremierBiosoft.com
NEWHELIX is a software program for the analysis of oligonucleotide structure. It can be found on the UCLA departmental server at URL http://www.mbi.ucla.edu.
OMIGA sequence analysis tool for Windows95 and NT. Performs multiple sequence alignments and contains over 50 DNA and protein analysis functions. Available from Accelrys.
Onto-Express automates the process of annotating a set of expressed genes with genomic functions. It quickly provides a graphic summary of genes by function including biological processes, biochemical function, molecular role, cellular role, cellular component, and chromosome location. The user uploads a set of expressed genes in either accession or cluster ID order. Once the resulting graph is displayed, the user can click on the function and see the list of genes that have that particular function in common. Clicking on a gene then beings up the UniGene database for further investigation. Version 1 is freely available over the web and can be accessed from www.openchannelfoundation.org. Version 2 adds in probability factors for each function so that the researcher can identify significant areas for further study. V2 is available by subscription.
Contact:
- Open Channel Software
- 1807 W. Sunnyside Ave, Suite 301
- Chicago, IL 60640
- Tel: (773) 334-8177 x 11
- Fax: (773) 334-8187
Dna Sequence Analysis software, free download. software
--- P, Q, R ---
PAP is a suite of programs to analyze protein structures. Modules are available to compute and display as a function of sequence Ramachandran and linear phi-psi plots, distance diagonal plots and temperature factors. Modules are also available to compute and plot RMS structure differences for two proteins. Proteins with sequence insertions or deletions can be handled. Priestle's RIBBON program has been converted to run as an interactive stand-alone program of SGI 4D series workstations. All programs accept Brookhaven PDB format for input and all programs have options to generate HP and PostScript output files. The programs run on Silicon Graphics 4D GT, GTX, Personal Iris and Indigo workstations. The programs were developed by T. Callahan, W.B. Gleason and T.P. Lybrand. It is available as program number 594 from QCPE
PatternHunter is a revolutionary general-purpose homology search tool based on well-recognized, innovative and proprietary technologies. It provides all the tools necessary for fast and sensitive homology search in all flavors, including: DNA-DNA, Protein-Protein, translated DNA-protein, and translated DNA-DNA searches. PatternHunter saves you time and money by delivering sequence comparisons with astonishing speed and accuracy.
Platforms: Platforms: All platforms supporting Java VM
Categories: Bioinformatics, Sequence analysis, Homology Search
For additional information, contact:
- Bioinformatics Solutions Inc.
- 145 Columbia St. West, Suite 2B
- Waterloo, ON Canada N2L 3L2
- Tel: (519) 885-8288
- Fax: (519) 885-9075
- http://www.bioinformaticssolutions.co m

PC/GENE was purchased by Oxford Molecular Group and is no longer available.
Dna Sequence Analysis software, free download Pdf
PEAKS is software that identifies proteins and peptides using tandem MS data. The PEAKS de novo approach is somewhat unique compared to other software like Mascot or Sequest. PEAKS uses a de novo sequencing component to generate peptide candidates. This is particularly useful for peptides not present in databases. Subsequently, a protein database searching component provides protein ID.PEAKS uses both the de novo results, and database search results to identify proteins.
Platforms: Windows, Linux, Java
Categories: Bioinformatics, Drug Discovery, Proteomics, Protein ID, De Novo Sequencing.
For additional information, contact:
- Bioinformatics Solutions Inc.
- 145 Columbia St. West, Suite 2B
- Waterloo, ON Canada N2L 3L2
- Tel: (519) 885-8288
- Fax: (519) 885-9075
- http://www.bioinformaticssolutions.co m
PepSeq is a web based training software designed for online computer training of peptide sequencing techniques to distance education and on-campus biochemistry students at university or college. It covers the concepts of alkaline hydrolyses (composition analysis), enzymatic and chemical fragmentations, and exopetidase digestions. Originally written as a DOS based program, PepSeq has been re-written as an online service which provides many benefits.
Plasmid is a new Windows-based vector map drawing and sequence analysis program that lets you:
- quickly and easily draw high resolution, publication quality circular or linear maps, with or without DNA sequences
- map restriction sites using an included tool set of nearly 1000 restriction enzymes.
- instantly convert sequence files on your computer or on the Internet (using Plasmid's integrated web browser) to fully labeled graphical maps.
- copy maps to the clipboard for pasting into other Windows applications such as Microsoft Word or PowerPoint.
Plasmid is available from Redasoft (http://www.redasoft.com)
PPNN from the LBNL, is a neural network program that will predict promoter regions.
PRIMER PREMIER 5: A fully featured, competitively priced primer design program to design specific, sensitive, high yield primers.
- Cross species primers - design primers for amplifying sequences from multiple species
- Pathogenic detection primers - design primers in highly conserved regions
- Allele specific primers - design primers to exclusively amplify a single member of a set of related sequences
- Degenerate primers - start with an amino acid sequence, reverse translate and design primers in regions of low degeneracy automatically
- Nested/multiplex primers - select primers for nested PCR reaction by checking multiple primer homology among candidate primers
- Restriction enzyme and motif analysis - select from the database of over 800 enzymes and 200 common
- ClustalW alignments - load the aligned sequences after performing Clustal W alignment using any resource, including EBI
- Multiple sequence alignment - use the popular Clustal W algorithm to align sequences from your desktop (available in Windows version only)
- Alignment editor - manually insert/delete/move gaps in an alignment
- Long PCR handles sequences up to 50 KB
Available for both Windows and Macintosh. PRIMER IER is available from:
PREMIER Biosoft International
3786 Corina Way
Palo Alto, CA 94303-4504
Phone: 650-856-2703
Fax: 650-843-1250
http://www.PremierBiosoft.com/
PROCHECK, from Thornton, et al., is a program which conducts knowledge-based analysis of protein structures. It will produce a geometric analysis of proteins and DNA including phi-psi angles, hbonding, secondary structure evaluation and other features of interest. The program is available from the Protein Data Bank
PROCRUSTES, from the University of Southern California uses the spliced alignment algorithm, to examine all possible exon assemblies and finds the multi-exon structure with the best fit to related protein.
Protein Networker enables an institute to make high throughput protein interaction data generated by two hybrid screens available throughout the institute. It generates graphical protein interaction maps for all protein interactions. It is a web based application allowing access on an intranet to any researcher with a browser. Secure logins ensure only authorized users have access. Secure access can optionally be enabled on the internet.
The database administrator is given additional tools for the addition and modification of interaction data. Addition of interaction data is provided by standard spreadsheet format files to ensure compatibility with other applications. Access for modification of the interaction data is restricted to authorized users. Available from
PREMIER Biosoft International
3786 Corina Way
Palo Alto, CA 94303-4504
Phone: 650-856-2703
Fax: 650-843-1250
http://www.PremierBiosoft.com
ProteinSolve is a single-User desktop sofware package for analyzing protein sequence infromation. The program runs on Microsoft Windows 98, NT, 2000; PowerMac, Solaris 2.6, Irix 6.2+ and Linux-x86. For additional information,
- Stratagene
- 11011 North Torrey Pines Road
- La Jolla, CA 92037 USA
- Tel: (800) 424-5444 or 800-894-1304
- Fax: (512) 321-3128
- Web: http://www.stratagene.com
The Rosetta Resolver System, provides high-capacity data storage, retrieval and analysis of gene expression data. The system is ideal for life science research organizations that need to assess compound specificity or toxicity, identify new genes or therapeutic targets, or compare and analyze large databases of expression profiles. The Rosetta Resolver system combines flexibility and ease of use with high-performance algorithms to produce an enterprise solution for rapid analysis of gene expression data. The system can accept and analyze data from a wide variety of expression profiling formats, and applies Rosetta's proprietary error models to yield quality statistics for every gene expression measurement. These statistics are automatically leveraged by all analysis tools, ensuring reliable results.
Additional new features of the Rosetta Resolver system version 2.0 include the following:
- Analysis of both intensity and ratio-based expression profiles
- Summarization and analysis of data at the feature, reporter (oligonucleotide, cDNA, or probe pair), sequence, exon, and UniGene cluster levels
- Automated sequence annotation updates from public and proprietary databases
- Ability to build ratio-based analyses from intensity-based analyses using advanced statistical error models that leverage replicate hybridizations
- GEML-compatibility enables users to export data in the GEML format for exchange with collaborators and/or for publication. GEML compatible software such as Rosetta's GEML Conductor tools can be used to visualize the data.
Version 2.0 is available now through Rosetta's strategic partner, Agilent Technologies. The system comes in several flexible packages to meet the needs of a variety of research organizations. Additional information about the Rosetta Resolver system can be found on Rosetta Inpharmatics' Web site at www.rii.com.
RGBG Analyzer software is designed for color analysis of digital images. It is intended for use by biomedical researchers for the determination of color values, including gray values, of various blots, such as western blots.
--- S, T, U ---
SEQtools 8.2 is a win32 software package for handling and analysis of nucleotide and protein sequences. The program includes a series of trivial functions to help you carry out common operations. In addition SEQtools will assist you with more demanding tasks like unattended batch blast search at NCBI. SEQtools includes advanced facilities for retrieving, storing, handling and listing search results. Special functions are included for design of microarray gene expression analysis experiments, for expression analyses with the SAGE procedure and for managing small EST projects. Utilities are included for primer design and ordering, renaming files, creating codon usage tables, building local searchable databases, aligning nucleotide and protein sequences, comparing sequences and a lot more... Additional information is available at the SEQtools website.
SIGNALSCAN, a service from the NIH, finds homologies in published signal sequences based on the sequence submitted. The program is an excellent method to learn the identity of unknown proteins bound to characterized binding sites in DNA sequences.
SimGlycan is a program for prediction of the theoretical glycan structures. This SimGlycan predicts the structure of a glycan from the MS/MS data acquired by mass spectrometry, facilitating glycosylation and post translational modification studies. SimGlycan accepts the experimental MS profiles generated by a mass spectrometer, matches them with its own database of theoretical fragmentation of over 7,000 glycans and generates a list of probable glycan structures. Each structure is scored to reflect how closely it matches your experimental data. Other biological information for the probable glycan structures such as the glycan class, reaction, pathway and enzyme are also made available for easy reference.
PREMIER Biosoft International
3786 Corina Way
Palo Alto, CA 94303-4504
Phone: 650-856-2703
Fax: 650-843-1250
http://www.PremierBiosoft.com
SimVector 4 is a web savvy program equipped with all the drawing tools necessary to publish engineered plasmid catalogs, as well as to simulate and draw plasmid constructs for Gateway®, TA, Restriction or other popular cloning techniques. With its powerful drawing features, it is easy to enhance vector maps with patterns, colors, fills, curved text and annotations. Available from
PREMIER Biosoft International
3786 Corina Way
Palo Alto, CA 94303-4504
Phone: 650-856-2703
Fax: 650-843-1250
http://www.PremierBiosoft.com
Solltech DNA fingerprinting and image analysis software, dendrogram capable. Contact
ALSBYTE Biotechnology Products
PO Box 339
Mill Valley, CA 94942-0339
ALSBYTE Biotechnology Products
2175 E. Francisco Blvd. Suite A4
San Rafael, CA 94901-5524
Tel: 415-457-8502
Fax: 415-457-8541
Email: alsbyte1@well.com
Web: http://www.alsbyte.com
STEM (The Short Time-series Expression Miner) is a Java program for clustering, comparing, and visualizing short time series gene expression data (8 time points or fewer). STEM allows researchers to identify significant temporal expression profiles and the genes associated with these profiles and to compare the behavior of these genes across multiple conditions. STEM is fully integrated with the Gene Ontology (GO) database and supports GO category gene enrichment analyses for sets of genes having the same temporal expression pattern. STEM also supports the ability to easily determine and visualize the behavior of genes belonging to a given GO category, identifying which temporal expression profiles were enriched for these genes.
STRAP is an editor for the multiple sequence alignment of proteins. The computer program STRAP supports the analysis of hundreds of proteins and integrates amino acid sequence, secondary structure, 3D-structure and genomic- and mRNA-sequence and residue annotation. The included tutorial will teach the use of STRAP in as little as one hour. The scriptability and extendability make STRAP a very powerful tool for even the most advanced users. The STRAP software is programmed in JAVA and runs on all computers. Strap is maintained and will be permanently improved.
TESS (Transcription Element Search Software), from the University of Pennsylvania, is a system for locating and displaying transcription factor binding sites in DNA sequences.
TurboBLAST is an accelerated, parallel implementation of BLAST, a sequence comparison tool that provides critical insight into the structure and function of genes and proteins. As submissions to sequence databases increase exponentially with the completion of large-scale sequencing projects around the world, the demand for BLAST processing has outstripped the IT resources of genomics and proteomics labs.
TurboBLAST delivers massive acceleration of BLAST by partitioning BLAST computations into manage-able tasks and distributing them across a network of workstations, PCs, or Macs. Initial benchmarks of TurboBLAST on a network of 11 commodity PCs running Linux reduced a month-long BLAST run to just two days. Greater speed-up of BLAST is achieved simply by adding more machines to the TurboBLAST system.
Based on Java technology, TurboBLAST runs on all types of computers, from commodity PCs and Macintosh computers to UNIX workstations and high-performance parallel supercomputers. Versions are available for most operating systems, including the most popular releases of Linux, Unix, Mac OS, and Windows. Contact
- Ed Pillsbury
- TurboGenomics
- One Century Tower
- 265 Church Street
- New Haven, CT 06510
- 203/974-0470 x222
- pillsbury@turbogenomics.com
- View and analysis of annotated DNA or protein sequences
- Chromatograms visualization and editing
- ORFs highlighting
- Integrated support of search requests to NCBI databases
- Multiple sequence alignment with optimized port of MUSCLE package
- A graphical interface for an optimized HMMER package
- 3D molecular protein and DNA structure viewer for PDB format
- Workflow Designer to build complex reusable computational tasks
- Search for Transcription Factor Binding Sites (TFBS) in DNA sequences
- Repeats analysis
- REBASE restriction enzymes database integration
- Huge files support, native support of gzipped documents
- Available for Windows, Linux and MacOS X platforms
UniPro DPview is a powerful tool for analyzing genetic sequences. It can be used for exact and inexact comparison of both DNA or protein fragments and whole chromosomes to find homologies and repeats, examine the matches found in an interactive mode, filter them using arbitrary criteria and create alignments based on this data. The program is available at http://bits.unipro.ru/eng/dpview.html.
--- V, W ---
Vector NTI Advance is the most highly integrated, multifunctional desktop sequence analysis application suite available today. It features a comprehensive set of data analysis and management tools, implemented across five application modules. All the modules share an information-rich graphical user interface that makes sequence analysis both simple and intuitive. Vector NTI Advance gives researchers the power to analyze proprietary sequences in the context of human genome project data. In addition, Vector NTI Advance contains an object-oriented database, accessible from all modules, that enables a wide range of cross-module bioinformatics workflows. No tedious data reformatting is necessary, and no proprietary file formats are created, so data import, export, sharing and collaboration are flexible and simple. Vector NTI Advance also contains tools to speed bench research, such as batch PCR primer design and in silico gel electrophoresis, and every representation of data within the suite is available for export to other applications for presentation purposes. For additional information contact:
- InforMax
- Invitrogen Life Science Software
- 7305 Executive Way
- Frederick, MD 21704
- Tel: 800-357-3114
- Fax: 240-379-4010
- Web Site: http://www.informaxinc.com
VIBE (Visual Integrated Bioinformatics Environment). Introducing a next generation visualization and genomics discovery tool: the Visual Integrated Bioinformatics Environment (VIBE). Developed by INCOGEN in collaboration with TimeLogic Corporation, VIBE is a visual data analysis and mining environment designed specifically for the TimeLogic DeCypher platform. This new, highly sophisticated client/server system, combined with TimeLogic's powerful DeCypher accelerators, offers the bioinformatics researcher an unprecedented total and integrated solution to data mining and informatics discovery.
Dna Sequence Analysis software, free download For Windows 10
- Efficient drag-and-drop sequence analysis pipeline construction
- Visual implementation of all DeCypher algorithms
- Data filtering on simple or complex criteria
- Distributed, multi-user support
- Interactive or batch mode pipeline execution
- User editable representation of pipelines in XML and Perl
- State-of-the-art, interactive visualization tools
- Intuitive, user-friendly data representation
- Results accessible from active and previously completed pipelines
System Recommendations: VIBE is a client/server system requiring Unix or Windows as the workstation operating system and Unix as the server-side operating system. A TimeLogic DeCypher server is also needed to execute the sequence analysis algorithms. Contact:
- Incogen, Inc.
- 263 McLaws Circle
- Suite 200
- Williamsburg, VA 23185
- Phone: 800-286-6599
- Fax: 864-654-0887
- E-mail: info@incogen.com
Visual Cloning 2000 is an advanced new vector map drawing and sequence analysis program that includes the following features:
- Sophisticated graphics and extensive customization of genetic maps, including multiple maps per pa
- Extensive web integration, including fast and easy conversion from a GenBank or EMBL file to a graphical map, and integrated online ordering of enzymes and prime
- Advanced restriction analysis, with over 1000 enzymes
- Open Reading Frames sear
- PCR primer and hybridization oligonucleotide desi
- Sequence view
- Subsequence sear
- Intuitive user interface
The beta version of Visual Cloning 2000 is available for download at http://www.redasoft.com.
Dna Sequence Analysis software, free download Torrent
The Wisconsin Package for sequence analysis contains over 100 interrelated software programs. It enables scientists to analyze DNA and protein sequences by editing, mapping, comparing, and aligning them. Other programs facilitate RNA secondary structure prediction, DNA fragment assembly, and evolutionary analysis. In addition, the major genetic databases--GenBank, EMBL, PIR, and SWISS-PROT--are distributed with the software and are fully accessible with the database searching and sequence manipulation programs. The Wisconsin Package runs on both OpenVMS and UNIX operating systems. And for those who want to modify or enhance the software, documented source code is available. The program is available from:
Accelrys
575 Science Drive
Madison, WI 53711
Tel: (714)955-2120
Fax: (714)955-2118
URL: http://www.accelrys.com